Claremont High School Life Computes: Lab 1
Today
This lab experiments with strings of DNA, RNA, and protein.You will use Python functions embedded in the website to model
- DNA replication
- DNA transcription into RNA
- RNA translation into proteins
DNA:
Concept Review: DNA replication vs. DNA transcription
There are two fundamental things that happen to DNA:- DNA replication (creating a copy of a cell's DNA)
- DNA transcription (creating a messenger RNA or mRNA)
Next, take a look at these two images. Label each with the process that it depicts.
Accessing the Site
To begin, open the site. You will need to log in with your user name and password. Once you are logged in, you will see a screen that will be blank other than three boxes located at the top. This current screen will eventually show any answers you submit so you can check your work. By hovering your mouse over a box, you will make links appear in a box below, like the below image. To access the functions you will need for today's lab, hover your mouse over the Simulations box, and click on the link labeled DNA Screen.
To access the functions you will need for today's lab, hover your mouse over the Simulations box, and click on the link labeled DNA Screen.
Visualizing DNA
You can visualize a string of DNA by typing it into the DNA text box. The visualization will automatically update whenever you click outside of the text box. IMAGE! of the beautiful DNA screen. You will see that the shape of each nucleotide tries to match the shapes in your textbook. As noted on the top of the DNA screen, capitalization does not matter (the code will automatically capitalize your input), and using any unexpected characters will result in blank spots. IMAGE before and afterChallenge 2: Which function is which?
You should have three buttons located below the DNA input box. These buttons will cause various transformations to your DNA string, and output the results to the screen.Function 1 Function 2 Function 3You can pass any DNA string to these mysterious functions by typing anything into the DNA box, then clicking any of the three buttons. Your task is to try out these functions and match them with one of three processes: DNA replication, DNA transcription, and inversion (going backwards). To figure out which is which, you should try each of them. See what the output is... and then see if you can tell whether fun1 is DNA replication, DNA transcription, and inversion (going backwards). As you determine what's what, enter your findings here:
Visualizing DNA and RNA strings
DNA transcription creates a strand of mRNA, messenger RNA. You can visualize both a DNA strand and its complementary mRNA by changing the selected option to "View DNA Transcription into RNA". screenshot RNA will not be displayed before your first start codon, nor will it be displayed after a stop codon! make sure screenshot above has nonsense before a tac and after a stop Use the chart located just above the DNA input box to find the start and stop codons. Codons will not be recognized before the start codon. This means you can have as much nonsense as you like before the first occurance of 'TAC' in your DNA. However, the stop codon will need to be a multiple of three characters away. Just because 'TACTACTTT' has the string 'ACT' (which corresponds to a stop codon) located in it, does not mean that this sequence has the stop codon in it! Try it yourself! Try deleting the second occurance of 'T' in this string, and see what happens!Problem 3: Reverse it! determining DNA from RNA
Some viruses, like HIV, contain RNA genetic material instead of DNA genetic material. To survive, those viruses use reverse transcriptase in order to create DNA from their own RNA. Note that- reverse indicates that the process is going backwards from the usual progression DNA-to-RNA
- transcrip indicates that it is transcsription that is running in reverse
- ase indicates that reverse transcriptase is an enzyme making this reaction occur
As a challenge, you'll act as reverse transcriptase! See if you can determine which strings of DNA will output the following two strings of RNA:
- UAGUCC
- GCUGAG
Translating from RNA to proteins
Translation is the process that decodes RNA to create proteins. Watch an animation of this in action:As you can see, the chain of amino acids "AA" together form a protein.
Now here is a codon matching chart. The amino acid names and their one-letter abbreviations are labelled on the perimeter of the circle-chart.
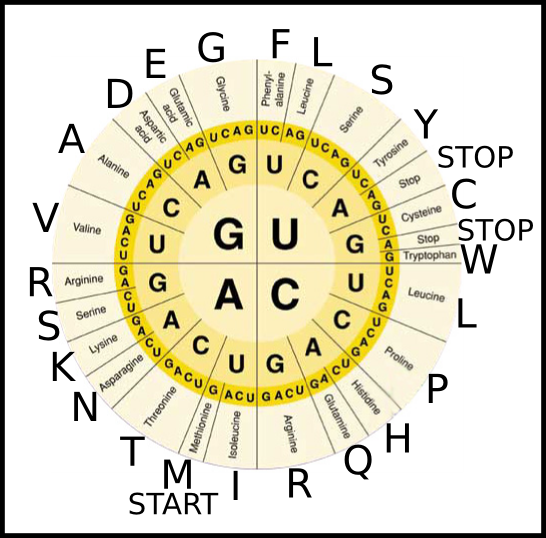
To get familiar with this chart and the one-letter abbreviations for amino acids, submit the amino acids that would be formed by the ribosome when it translates the following strings of RNA. Use the single-letter representations from the genetic code chart above. The first one is already solved as an example. To go from RNA to Amino Acid using the chart, you will need to work from the center to the outside. First find the quadrant of the chart with "G" on the innermost ring. On the next ring within that quadrant, look for the next letter, in this case, "A". On the outer most layer, then look for "C". You should be at "Aspartic Acid" now, which is designated as "D".
Problem 4: Lost in translation
In your life you have experienced good translations - and bad ones. By testing the three functions trans1( rna ), trans2( rna ), and trans3( rna ), determine which one of the three is the good translation function. The other two are incorrect. All three of these functions take an RNA string as input, and all three output a protein -- but only one of them outputs the correct protein! To get you started, you might try testing single codons - for example,>>> trans1( 'AUG' )See what the answer is, and then see if that answer is correct... . Be sure to check all three possible functions. Hint: you will need to try strings of RNA with two codons in order to answer this! Submit the correct translation function here:
Visualizing the process of transcription and translation (DNA to RNA to proteins)
You can use the "View RNA Translation into Amino Acids" option to visualize all three steps in biology's information change.- At the bottom, it will show the input DNA string.
- Above it, it will show the transcribed RNA string.
- Above that, you will see the Amino Acids produced by translation.
Problem 5: Protein Words
This "View RNA Translation into Amino Acids" function will help you connect DNA, RNA, and protein strings. Here, you will build words using the single-letter amino acids from the above codon chart showing the genetic code. For this problem, as in real life, you must begin the sequence with the START codon in DNA: TAC. You also must end with a STOP codon, either ATT, ATC, or ACT. For example, in order to build the protein word MEET, you could use the codons- TAC (the codon for 'M' and the start codon)
- CTC (the codon for 'E')
- CTC (the codon for 'E')
- ??? (the codon for 'T')
- ATT (one of the STOP codons)
Biology's central dogma
Biology stores, retrieves, and processes information -- just as computers do. The difference is that biological information is stored in DNA's genes. To retrieve it, living things- first, transcribe DNA to mRNA
- second, translate mRNA to proteins
- third, use the proteins to create cells of different types
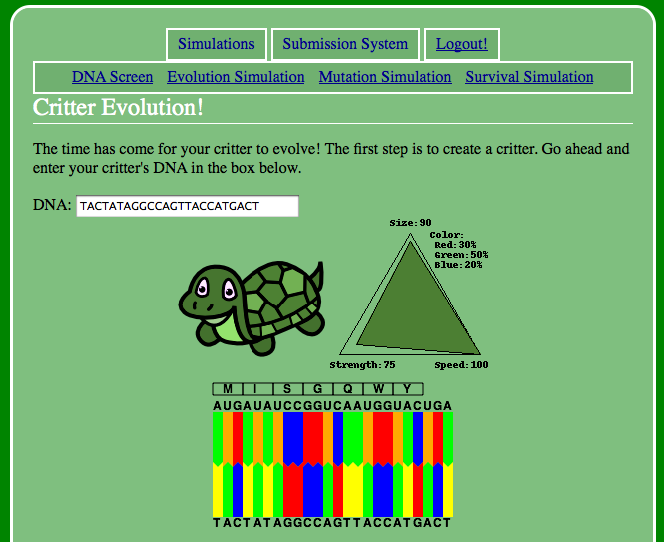 The previous DNA Screen was a lot more lenient than this new Critter Evolution screen. Notice that if you have any invalid characters at all in the new DNA box, the turtle will be unable to survive! Additionally, if the number of amino acids in its protein are not exactly 7 (including the start codon, excluding the stop), then the turtle will die.
By changing the codons, you will be able to change the various attributes of the turtle. Try seeing if you can figure out how to change the turtle's color, and if you can make the turtle smaller.
As a hint, the first three codons affect the color of the critter. Can you tell how?
The final three codons affect the strength, speed, and size. Can you tell which is which?
In the next lab, we will examine how mutations in DNA yield mutations in proteins, which produce different critters...
The previous DNA Screen was a lot more lenient than this new Critter Evolution screen. Notice that if you have any invalid characters at all in the new DNA box, the turtle will be unable to survive! Additionally, if the number of amino acids in its protein are not exactly 7 (including the start codon, excluding the stop), then the turtle will die.
By changing the codons, you will be able to change the various attributes of the turtle. Try seeing if you can figure out how to change the turtle's color, and if you can make the turtle smaller.
As a hint, the first three codons affect the color of the critter. Can you tell how?
The final three codons affect the strength, speed, and size. Can you tell which is which?
In the next lab, we will examine how mutations in DNA yield mutations in proteins, which produce different critters...
Extra: Behind the Scenes! How do we make a computer go from DNA to Amino Acids?
You should now have a rough understanding of how Amino Acid chains are created from DNA strands in biological systems. But what about in simulations such as this? How do you get a computer to understand what a DNA strand could mean? Computers are actually very similar to people in how they solve problems. If you ask it to solve a problem, but don't phrase the problem understandably, it can't solve it! Additionally, it can't solve problems unless you teach it how to first (generally by writing functions). So say we first start with the problem: "Computer, I'm going to give you a string* of A, T, C, and G. This string could be any length, I might forget to capitalize the letters, and there's a chance I'll make a mistake and give you a letter that doesn't correspond to a nucleotide! I want you to tell me what letters representing Amino Acids this strand corresponds to!" * a string is a data type that's basically a list of characters. Strings are convenient because they are entered in a very legible manner, IE: 'TACGAT' is a string. There are also easy ways for us to get only certain letters out of a string, and to add letters to a string, which will come in handy later. If you turn to someone sitting next to you and ask them to do this, you can imagine that they'd do something along the following:- Find some chart that goes from 3 nucleotide bases into amino acids (like the one above!)
- "Slice" or seperate your DNA string into 3 nucleotide chunks.
- Go through slice by slice, matching them against the chart, writing down what letter Amino Acid it should be
- Give you back the new string of letters.
>>> codonToAA('TAC')
'M'
The first line is what instructions we'd give the computer (the >>> is the start of the prompt), and the second is the output: the string 'M'.
This function would essentially be the same thing as your friend looking up in a chart what Amino Acid a codon refers to.
But wait! This codonToAA function would be useless for something like 'TACGAT', since the chart only goes 1 codon at a time! We'll need to slice the DNA to be useful.
If we want the first three characters out of this string, we can do
>>> dnaString = 'TACGAT' >>> dnaString[0:3] 'TAC'The first line makes the variable dnaString refer to our string! (Yes, this is just like variables in Math, such as x = 2) The second essentially says "Go from the start (the 0th position, if you will) of the string, and stop just before you hit the 3rd** position". ** The first character of a string is at position 0, NOT position 1! So if we wanted to translate this DNA string, its as simple as us doing
def dnaToAA(dna): return codonToAA(dna[0:3]) + codonToAA(dna[3:6])We're now beginning to define this as new function. The second function does exactly what it looks like. It returns (or outputs) the results of our "Look up in the chart" function after we pass it the slices. But wait, what happens if our input DNA isn't exactly 6 characters? If its not at least 6 characters, then our second slice will try to catch information that doesn't exist! If its more than 6 characters, we're not going to look up what that information means! You could imagine copy and pasting the codonToAA function over and over, making it take different slices. But this would be problematic. How does the computer know when to stop? How do you ensure that we don't forget to look up something? This is where the idea of Recursion comes in. Image: Recursion, did you mean Recursion? Recursion is a problem solving technique. If we were to try instead
def dnaToAA(dna): return codonToAA(dna[0:3]) + dnaToAA(dna[3:])The second line now tells our function to repeat the process. Yes, you can make functions that repeat themselves! You might notice that our second slice now doesn't tell python where its supposed to end. In this case, it will default to the end of the string. So basically, the second part of the second line says "Repeat this function, but remove the first three characters, that way you can look at the next codon!" This solves our problem of not knowing how many times we're supposed to repeat looking up in the chart. But now we hit another problem! How does this function know when to stop? Indeed, it now has no idea when its done! Logically, you know when to stop using the chart, right? If your codon is only 2 nucleotides, or if there aren't any at all, it doesn't make sense to use the chart! At this point, you're also probably at the end of the string, and thus done! So we add this "base condition" to the function.
def dnaToAA(dna):
if len(dna) < 3:
return ''
else:
return codonToAA(dna[0:3]) + dnaToAA(dna[3:])
The new if statement at the top works exactly how you think it would: If it sees that the length of the DNA is less than three, it'll do the indented next line, in this case, output a string with nothing in it and (more importantly) stop looping!
And with these 5 lines of code, we now have a function that can turn any string of DNA into its associated amino acids! The power of recursion makes it so we don't need to know how long the DNA is.
Recursion can be used to solve a wide variety of problems, as long as there are two things. - 1) A stopping point, or "Base Case" where the function knows its finished once its hit that point
- 2) The problem is doable by repeating a certain process over and over.
- I have a backpack that has a certain amount of space. Each item has its own size and how heavy it is. I want to pack the backpack so that its full, but as light as possible. What is the minimum weight of the stuff?
- I want to go from Los Angeles to New York by airplane. I can take either a direct flight, or I can make connections at several airports to eventually get there. What's the minimum price for flying to New York?
Extra Challenge: Recursion in action -> action in Recursion
One of the better ways to understand how to do recursion is to do recursion. Thus, this challenge is to follow the Tautology's instructions, and write a recursive function! You are going to want either IDLE or a Terminal window open. If you choose to do this via the Terminal's commandline, simply type "python" or "IDLE" to start up the shell. You can type in commands now! Try this quickly with simple commands such as>>> 5*5 25Now, when you are ready, type the following commands line by line into the shell. You will need to press tab as appropriate.
def UP(stuff):
if len(stuff) == 0:
return ""
else:
return stuff[0].upper() + UP(stuff[1:])
The first four lines should all look familiar. We're defining a function, then setting a base case.
The last line might look a tad unfamiliar. stuff[0] is how Python grabs the first character of a string. The upper function does just what it sounds like. The very last bit should be familiar though, removing the first character, then repeating the function.
Now try something like >>> UP("Please use Your besT Handwriting ON this") . Try any string you can think of! (Remember: when entering a string into Python, you MUST put quotes around it. Either single or double, but be consistent)
-- RaiFeren - 05 Aug 2010